Comparison between spatial regression methods
Comparison
library(agridat)
library(sf)
## Linking to GEOS 3.11.2, GDAL 3.6.2, PROJ 9.2.0; sf_use_s2() is TRUE
library(tidyverse)
## ── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
## ✔ dplyr 1.1.2 ✔ readr 2.1.4
## ✔ forcats 1.0.0 ✔ stringr 1.5.0
## ✔ ggplot2 3.4.3 ✔ tibble 3.2.1
## ✔ lubridate 1.9.2 ✔ tidyr 1.3.0
## ✔ purrr 1.0.2
## ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
## ✖ dplyr::filter() masks stats::filter()
## ✖ dplyr::lag() masks stats::lag()
## ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
library(spdep)
## Loading required package: spData
## The legacy packages maptools, rgdal, and rgeos, underpinning the sp package,
## which was just loaded, will retire in October 2023.
## Please refer to R-spatial evolution reports for details, especially
## https://r-spatial.org/r/2023/05/15/evolution4.html.
## It may be desirable to make the sf package available;
## package maintainers should consider adding sf to Suggests:.
## The sp package is now running under evolution status 2
## (status 2 uses the sf package in place of rgdal)
## To access larger datasets in this package, install the spDataLarge
## package with: `install.packages('spDataLarge',
## repos='https://nowosad.github.io/drat/', type='source')`
library(spatialreg)
## Loading required package: Matrix
##
## Attaching package: 'Matrix'
##
## The following objects are masked from 'package:tidyr':
##
## expand, pack, unpack
##
##
## Attaching package: 'spatialreg'
##
## The following objects are masked from 'package:spdep':
##
## get.ClusterOption, get.coresOption, get.mcOption,
## get.VerboseOption, get.ZeroPolicyOption, set.ClusterOption,
## set.coresOption, set.mcOption, set.VerboseOption,
## set.ZeroPolicyOption
data("lasrosas.corn")
yield_map <- lasrosas.corn %>%
sf::st_as_sf(coords = c("long","lat"))
yield_map_1 <- yield_map %>%
filter(year==1999)
yield_map_2 <- yield_map %>%
filter(year==1999)
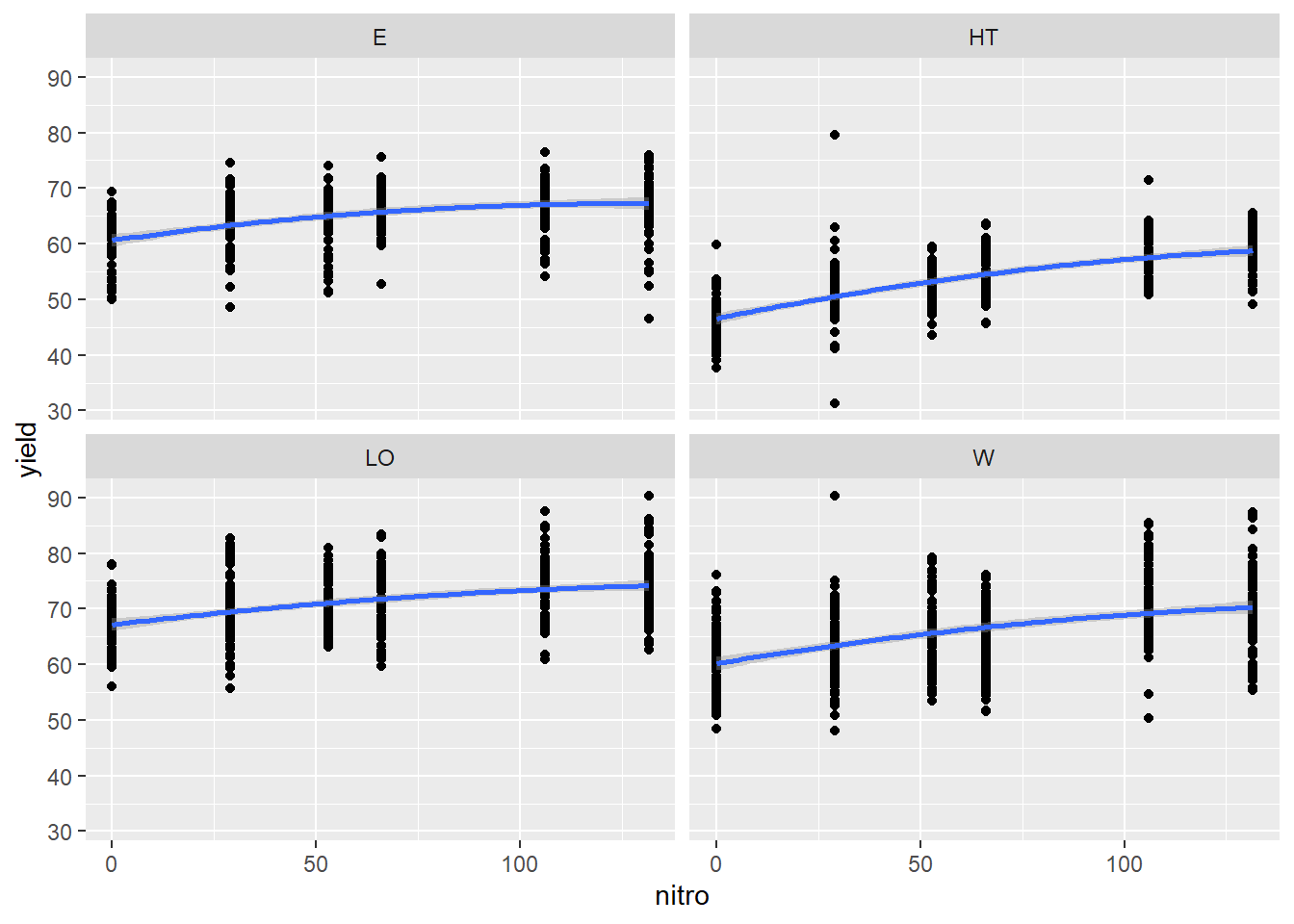
ggplot(data = yield_map_1,aes(x=nitro,y=yield)) +
geom_point() +
geom_smooth(method = "lm",formula = y~ 1 + x + I(x^2)) +
facet_wrap(~topo)
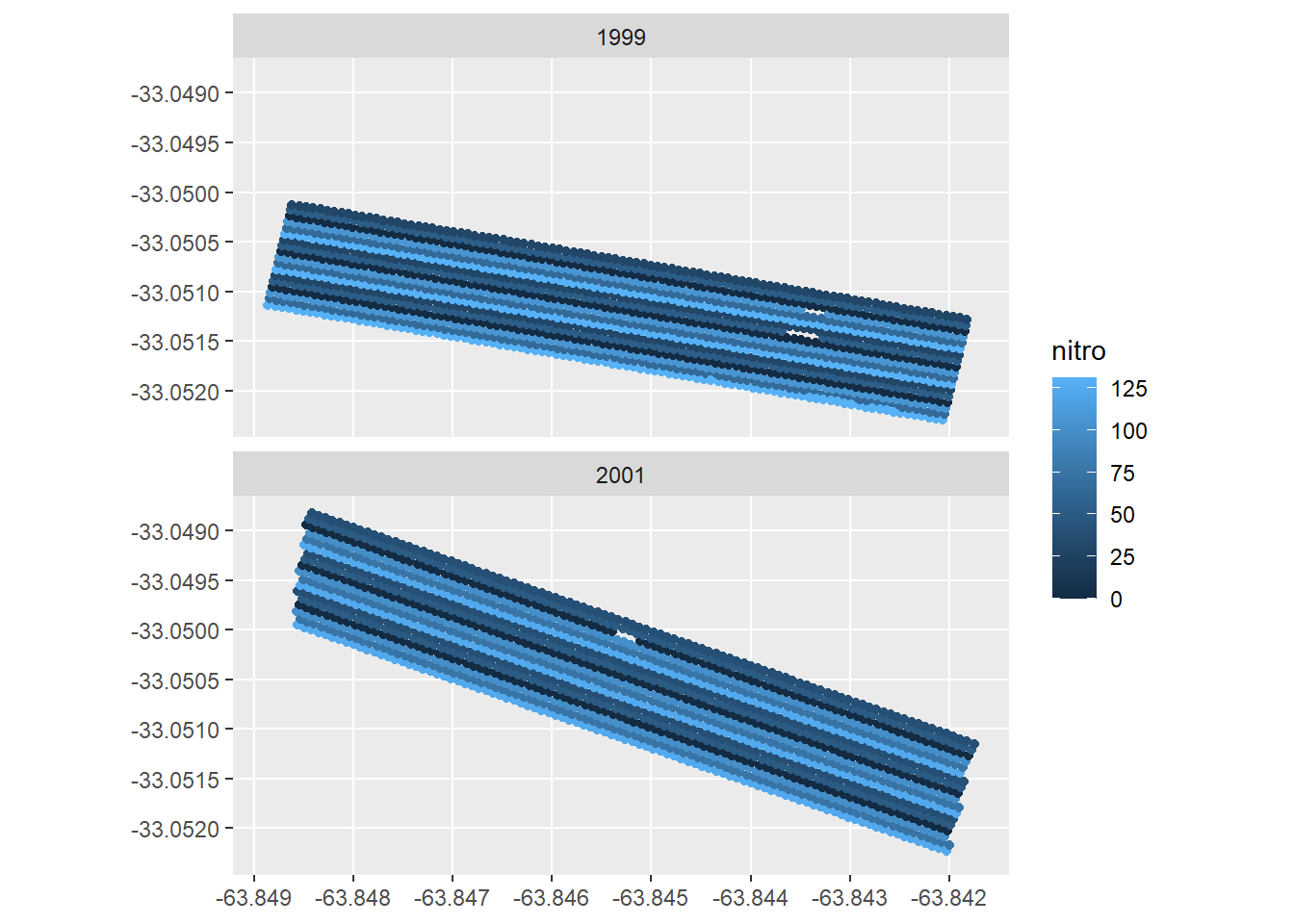
ggplot(data = yield_map) +
geom_sf(aes(color=nitro )) +
facet_wrap(~year,ncol = 1)
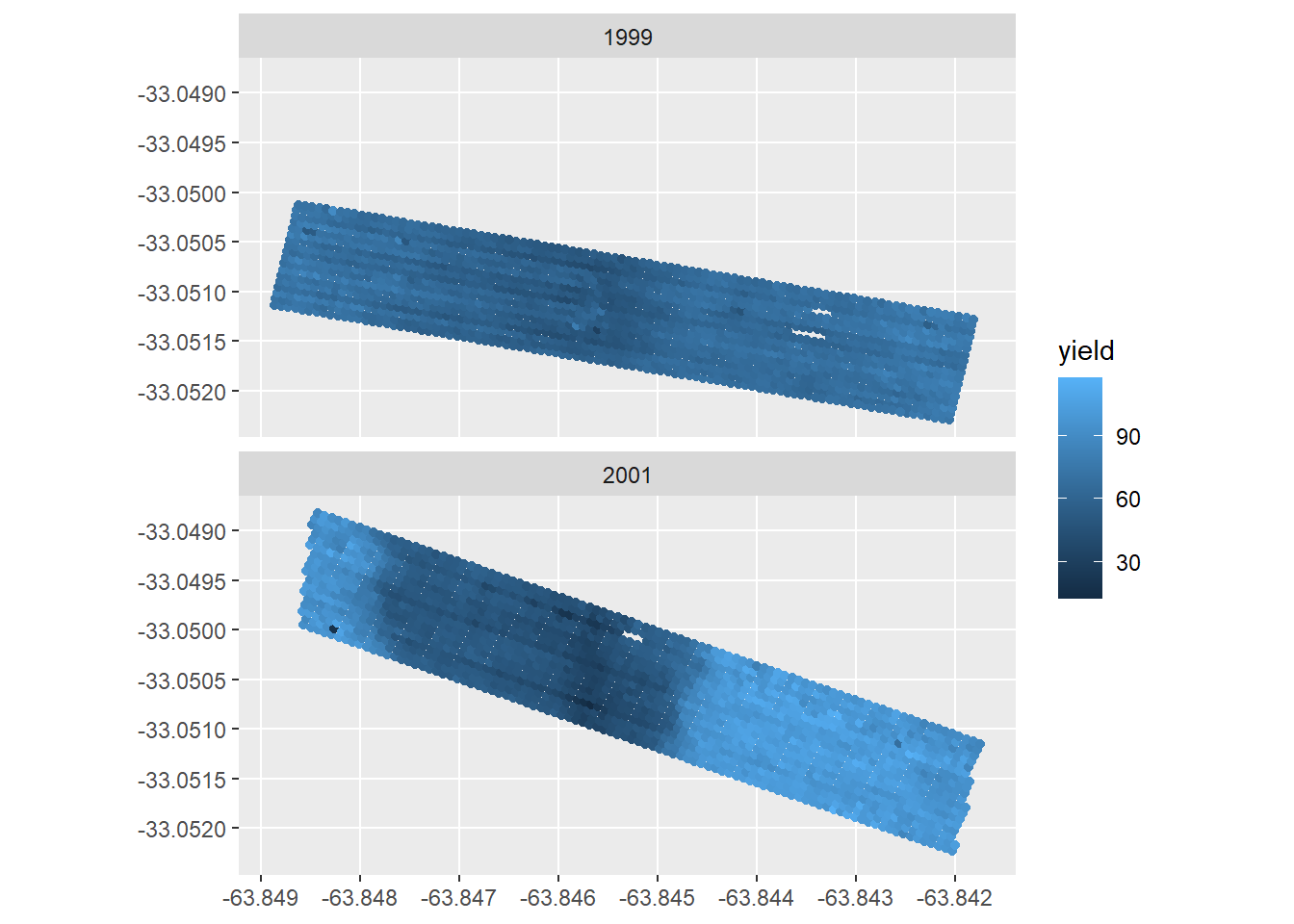
ggplot(data = yield_map) +
geom_sf(aes(color=yield )) +
facet_wrap(~year,ncol = 1)
# Modifying the dataset
trials <- yield_map %>%
mutate(N = nitro,
N2 = (nitro^2))
# Creating the W matrix
lw <- trials %>%
st_set_crs(st_crs("epsg:4326")) %>%
st_transform(crs=3744) %>%
st_centroid() %>%
dplyr::mutate(X = sf::st_coordinates(.)[,1],
Y = sf::st_coordinates(.)[,2]) %>%
st_coordinates() %>%
dnearneigh(0,35) %>%
nb2listw(style = "W")
bayesian_spatial_reg <- spBreg_err(formula = yield ~ N + N2,
data = trials,
listw = lw,
control = c(
ndraw = 50000L, # iteration
nomit = 15000L, # Burn in
thin = 5L, # thinning rate
prior = list(sige = list(nu = 0.01, d0 = 0.01), # informative Gamma(nu,d0) prior on sige (residual variance)
rho = list(a1 = 2, a2 = 2), # parameter for beta(a1,a2) prior on rho
lambda = 0.5,
Tbeta = diag(5) * 1000)))
## Warning in spBreg_err(formula = yield ~ N + N2, data = trials, listw = lw, :
## unknown names in control: prior.sige, prior.rho, prior.lambda, prior.Tbeta
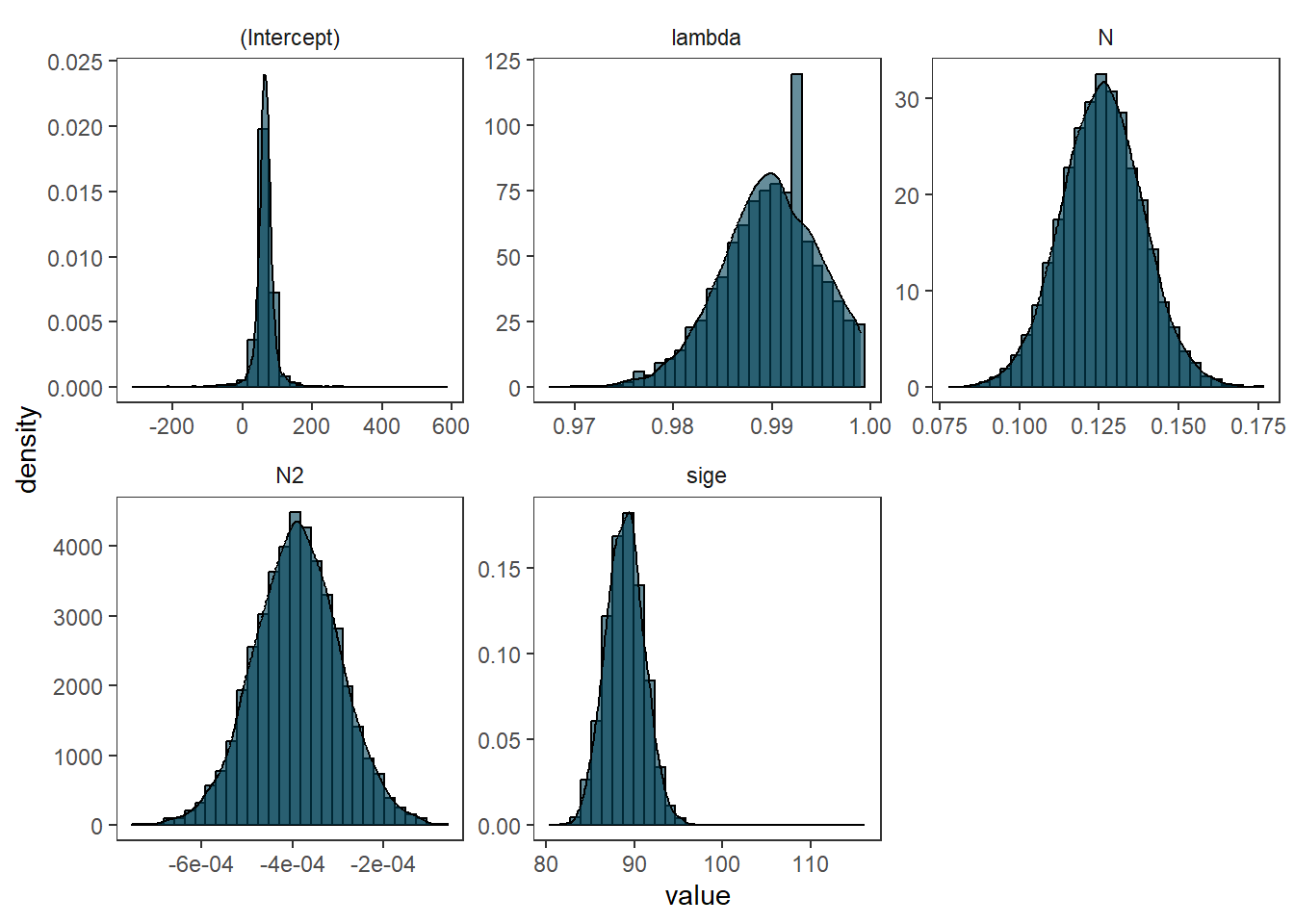
as.data.frame(bayesian_spatial_reg) %>%
pivot_longer(cols = everything()) %>%
ggplot(aes(value))+
geom_histogram(aes(y = ..density..), colour = 1, fill = "#004156", alpha =.6)+
geom_density(fill = "#004156", alpha = .6)+
facet_wrap(~name, scales = 'free')+
theme_bw()+
theme(strip.background = element_blank(),
panel.grid = element_blank())
## Warning: The dot-dot notation (`..density..`) was deprecated in ggplot2 3.4.0.
## ℹ Please use `after_stat(density)` instead.
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
## generated.
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.